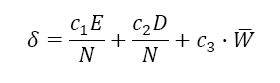NeuroEvolution of Augmenting Topologies
NEAT is an efficient system for evolving artificial neural networks in a genetic algorithm. NEAT can evolve extremely complex minimalist solutions to a variety of problems. NEAT is also one of the few neuroevolutionary systems that can perform evolution using both mutation and crossover operators. It is not always obvious how to perform a crossover operation in a neural network because the structures of different neural networks are not necessarily related. Therefore crossover operators have to perform complex analysis of the network structure to find appropriate points for crossover of the neural networks or phenomes. This problem is compounded by the fact that the genomic representation of the phenome does not clearly indicate where crossover can and cannot occur.
NEAT solves this problem by using historical markings in each element of each genome. Each node and link in NEAT added after the initial population of genomes is created has a historical marking attached to it. This guarantees that new innovations in structure are identifiable and recorded. This is just one of the ways in which NEAT is an innovative neuroevolutionary algorithm.
Genetic Encoding for NEAT
This section explains how genetic encoding works in NEAT. The genetic encoding for NEAT is slightly more complex than for some other neuroevolutionary algorithms simply because a NEAT gene encodes more information.
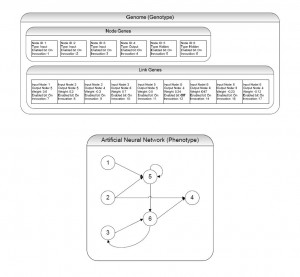
Each genome in NEAT contains a list of links and a list of nodes. Each link and node in these lists is referred to as a gene of the genome, and a genome comprises the entire set of hereditary information for an individual. Each link contains values for its input node and output node, the connection weight, information about whether the link is enabled or disabled and an innovation number. The innovation number serves as the link’s historical marking, denoting hereditary information about the gene. The innovation number allows the crossover algorithm to detect if its gene is similar to another innovation in a different genome. A node contains slightly less information than a link, including a unique node identification number, an activation response value, a disable bit variable and an innovation number. When the genome is converted to its neural network manifestation, it is referred to as a phenome. A phenome is the virtually-physical interpretation of the genotypes of the genome. Proper phenome structure must be conserved stringently during mutation and crossover operations. The link and node connectivity information comprise the physical characteristic, or genotype, information of the genome. When the genotype information is expressed physically, the observable characteristics are called the phenotypes.
Figure 1: Genotype and Phenotype Example for the NEAT Algorithm. Above is an example of a genotype that represents the displayed phenotype. There are six nodes: three input, two hidden and one output. There are nine links, two of which are recurrent and one of which is disabled. The disabled gene (connecting nodes 5 and 4) is not displayed.
NEAT mutation operations
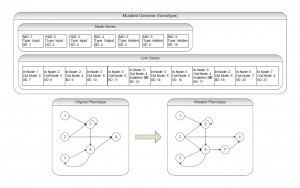
Mutation operations in NEAT can change connection weights, node activation values and network topology. All these mutations occur randomly, constrained by a fixed probability which is defined for each individual simulation. Weight and activation value mutations occur when a random node or link is chosen and its weight is perturbed by a constrained random value. NEAT mutation operations change network topology by adding links and nodes. When the GA mutates a link, it randomly chooses two nodes and inserts a new link gene with an initial weight of one. If a link already existed between the chosen nodes but was disabled, the GA re-enables it. Finally if there is no link between the chosen nodes and an equivalent link has already been created by another genome in this population this link is created with the same innovation number as the previously created link as it is not a newly emergent innovation. A node mutation is similar to a link mutation but differs from it in that instead of choosing two nodes and inserting a link, the GA chooses and disables an existing link and inserts a node. The GA inserts this new node with a random activation value, as well as two link genes to connect the node to the now-disabled link’s previous input and output nodes. The GA then transfers the weight from the disabled link gene to the new link gene, which is connected to the old output neuron. The weight of the link gene inserted between the new neuron and the old input node is set to one so as not to disturb any learning that has already occurred in this connection. Introducing a new node where a link once existed may fragment some evolved knowledge in the phenome. Copying the original link weight to one of the new node’s links while setting the other connecting link weight to one minimizes the disturbance in learning.
Figure 2: Mutation Example: A mutated form of the original genome displayed in figure 3.1 is shown here. Both a link and a node mutation have occurred to create a new phenotype. A link genome has been added to connect node 2 and 6. During the node mutation the link between node 6 and 4 was disabled and a new node (node 7) was added. In the definition of the link genes, the values once contained in the now-disabled link have been added to the link between nodes 7 and 4 and the link between nodes 6 and 7 has been set to a value of one to preserve the original learned value.
These mutation functions introduce complexity into the initial population, referred to as base genomes, and gradually grow a solution to the given fitness function. Since the processes are pseudo random a diverse population of genomes will evolve. The crossover function must be able to recombine these inherently different topologies efficiently. It does this using historical markings also known as innovation numbers.
Because the innovation numbers are unique to innovations and not to genes, it is possible to compare any two genomes in the population and determine which genes they share. If two genes share the same innovation number they also share the same manifestation, or phenotype. Innovations are preserved among genomes in the same population. In order for mutation and crossover to function in NEAT, the system must maintain a database of all the innovations that have occurred since the first generation of each simulation. When a new innovation occurs it is checked against the database of innovations to ensure that it is not identical to an existing innovation. If its originality is confirmed, a global innovation number is incremented and assigned to it and it is recorded in the innovation database. This guarantees that while each genome might have a different structure with different weights, all related genes are identical. When a crossover operator is applied to two genomes the offspring inherits the same innovation number in each gene. This preserves the historical markings through generations. This preservation of historical markings prevents the crossover operation from becoming too computationally intensive and the networks from exploding in size because of crossover.
Crossover in NEAT
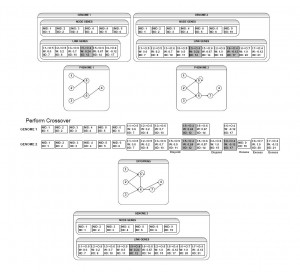
Figure 3: Crossover Operation: Although the parents are structurally different, their innovation numbers show that they are very closely related. Crossover happens easily without requiring structural analysis.
During a crossover operation, NEAT can quickly determine how to line up the two parents’ genes. Once they are aligned it is easy to see which portions are similar and which are different. Any genes that do not share innovation numbers with genes in the other parent’s genome are referred to as disjoint and are added to the child during crossover. If either parent has genes that are newer than any of the genes in the other parent, they are considered excess and are also added to the child during crossover. All genes that are shared by both parents are inherited by the child from the parent with the highest fitness. A gene that is disabled in one parent but enabled the other has a chance of being re-enabled in the offspring.
This method of crossover allows NEAT to build increasingly complex ANN structures without restricting compatibility between genomes. Unfortunately this level of complexity works against the genetic algorithm as it cannot support such diversity in its population. A structural innovation that could exceptionally improve performance in a later generation may introduce a major change in a given genome, but since it requires a few generations to reach its full potential it may be erased from the population before it has a chance to affect performance. This is why NEAT employs speciation: to protect genomic innovation.
Speciation
In natural evolution entities that once shared a common genome sometimes diverge so much that they can no longer mate with one another. This divergence is known as speciation. In NEAT, as the genomes in a population grow complexity a new innovation in their topology may result in greater performance for the population’s agents. NEAT uses speciation to protect such innovations. When an agent’s structure diverges far enough from that of the other agents in the population NEAT identifies it and places it in its own species. Using innovation numbers NEAT can calculate the distance between two genomes. The distance is defined by the following function:
Figure 4: Distance Between Genomes: The distance d between two genomes is the sum of the number of excess (E) and disjoint (D) genes, and the average of the weight differences of the two genomes (W ¯). The coefficients c1, c2, and c3 modify the weights of each of the variables and N is the number of genes in the larger of the two genomes.
Initially one species is formed from the entire first generation. The first genome in the generation becomes the champion of that species since the population is uniform. As the algorithm proceeds and more complexity is introduced distances between genomes will increase until they are larger than the distance threshold. At this point, NEAT designates this structurally different genome a new species and names it as the species champion. As other genomes’ distances from their species champion increases, they may be placed in a different existing species if their distance from that species champion becomes small enough.
NEAT maintains species through generations to protect innovation and as an evaluation method for the effectiveness of an innovation. If no members of a species rise above their existing champion in fitness for a set number of generations, the entire species is terminated, unless its champion is the population champion.
To determine the number of genomes each species can introduce into the next generation, NEAT uses explicit fitness sharing. Each species is assigned a certain number of reproduction spots based on the sum of the species’ adjusted fitness values. Each genome’s adjusted fitness score is based on its distance from every other genome in the population. The lowest-performing fraction of each species does not reproduce, and the highest performer from each species carries over to the next generation via per-species elitism. Any remaining reproduction spots are filled through random selection.
If a species becomes too large, its genomes cannot reproduce productively because they do not have enough reproduction spots in the next genome. This keeps the species’ sizes reasonable and is necessary for speciation-based evolution systems. If species size were not restricted one species could grow to dominate the entire population and the benefit of speciation would be lost. Most genomes in a species are structurally similar because new structural innovations are slowly added to the phenotypes, reducing the generation by generation structural variation. Hence speciation protects innovation.
The NEAT algorithm is a robust EANN. It uses speciation to protect innovation, and it uses innovation numbers to perform all GA operations efficiently. The efficiency of the GA operators helps NEAT limit increasing complexity. This combination allows NEAT to search a broad solution space efficiently while minimizing the complexity of its solutions.